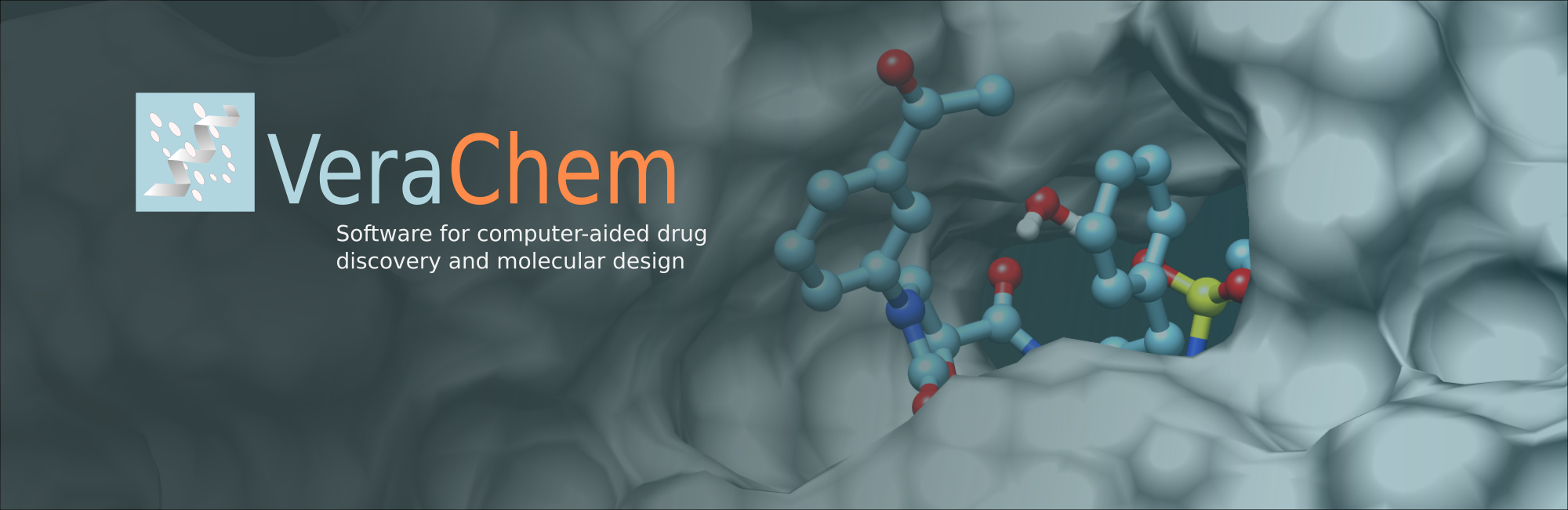
A joint VeraChem, Iowa State University, and University of Colorado Denver study applying QM-VM2 to the calculation of host-guest binding free energies published in the Journal of Chemical Physics.
QM-VM2, which efficiently combines statistical mechanics with quantum mechanical (QM) energy potentials in order to calculate noncovalent binding free energies of host–guest systems, is presented. QM-VM2 efficiently couples the use of semi-empirical QM (SEQM) energies and geometry optimizations with an underlying molecular mechanics (MM) based conformational search, to find low...